The plot is created based on a grid (rows, cols). Each grid point is numbered from bottom to
top and left to right (starting from 1), i.e. given grid point with coordinates (r, c) (where
(1,1) is the top left corner and (rows, cols) is the bottom right corner) the grid id is `(c
rows + r`. You must assign a node to the hypergraph to a grid point (see below).
Usage
plotHypergraph(
hgf,
gridDim,
showGrid = FALSE,
radx = 0.03,
rady = 0.05,
cex = 1,
marX = 0.035,
marY = 0.15,
...
)Arguments
- hgf
A list with the hypergraph containing two data frames, normally found using
getHypergraph(). The data framenodesmust have columns:sId(state id),gId(grid id) andlabel(node label). The data framehyperarcsmust have columnssId(head node),trans<n>(tail nodes),aIdx(action index),label(action label),lwd(hyperarc line width),lty(hyperarc line type) andcol(hyperarc color).- gridDim
A 2-dim vector (rows, cols) representing the size of the grid.
- showGrid
If true show the grid points (good for debugging).
- radx
Horizontal radius of the box.
- rady
Vertical radius of the box.
- cex
Relative size of text.
- marX
Horizontal margin.
- marY
Vertical margin.
- ...
Graphical parameters passed to
textempty.
Examples
## Set working dir
wd <- setwd(system.file("models", package = "MDP2"))
#### A finite-horizon replacement problem ####
mdp<-loadMDP("machine1_")
#> Read binary files (0.000124503 sec.)
#> Build the HMDP (3.4901e-05 sec.)
#> Checking MDP and found no errors (1.5e-06 sec.)
plot(mdp)
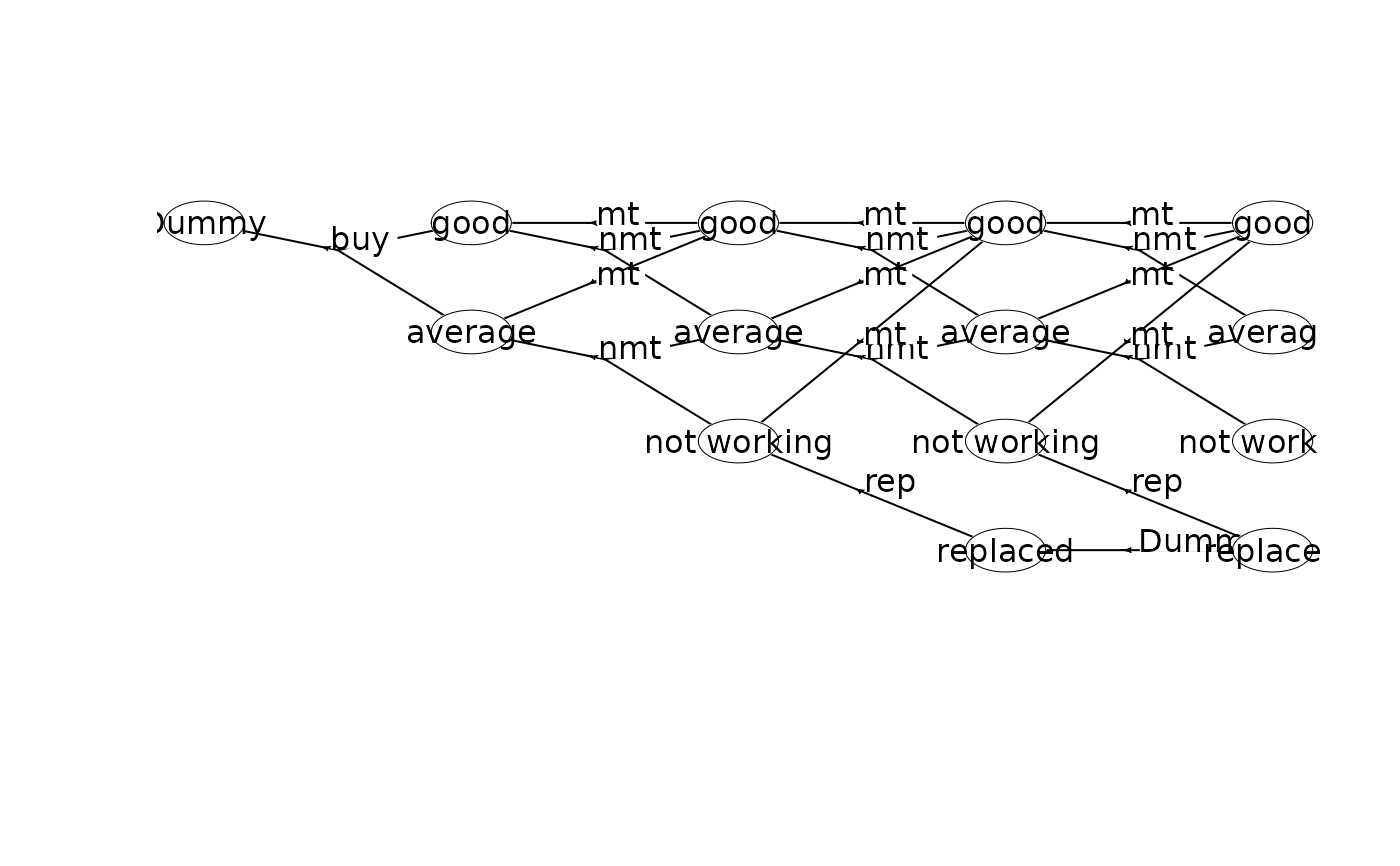 plot(mdp, hyperarcColor = "label") # colors based on labels
plot(mdp, hyperarcColor = "label") # colors based on labels
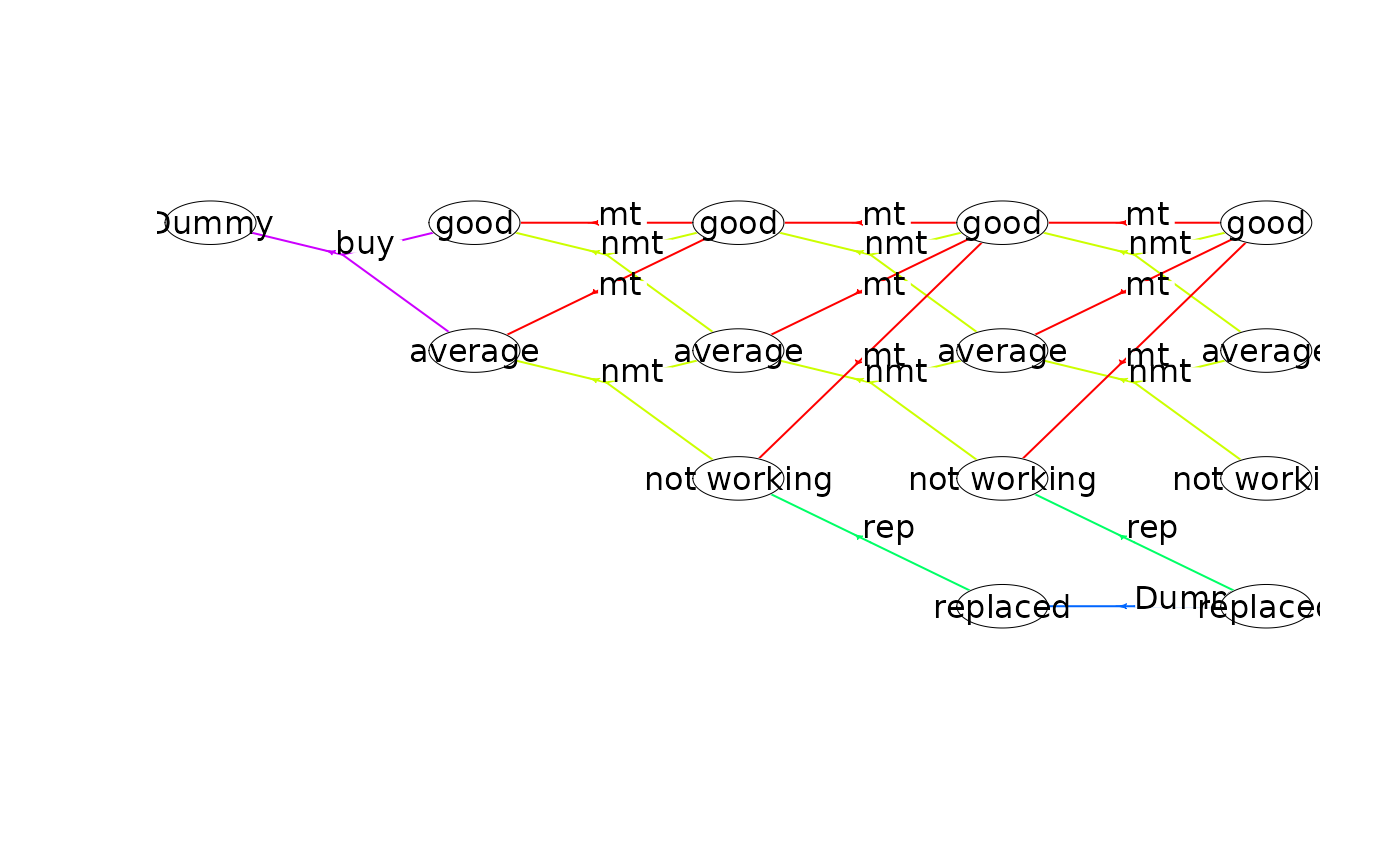 plot(mdp, hyperarcColor = "label", nodeLabel = "sId:label") # node labels are 'sId: label'
plot(mdp, hyperarcColor = "label", nodeLabel = "sId:label") # node labels are 'sId: label'
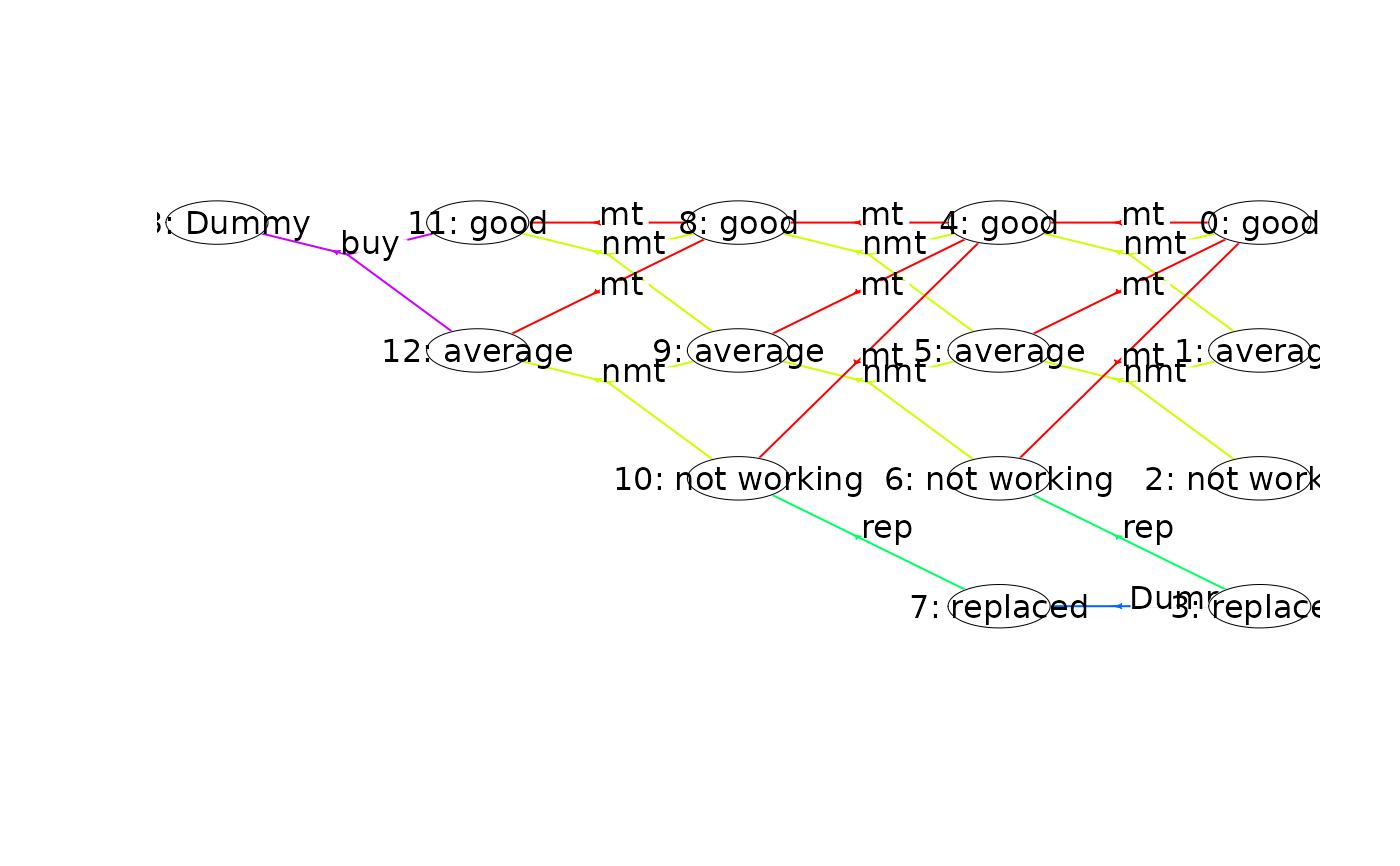 plot(mdp, nodeLabel = "sIdx:label", radx = 0.02) # adjust radx in nodes
plot(mdp, nodeLabel = "sIdx:label", radx = 0.02) # adjust radx in nodes
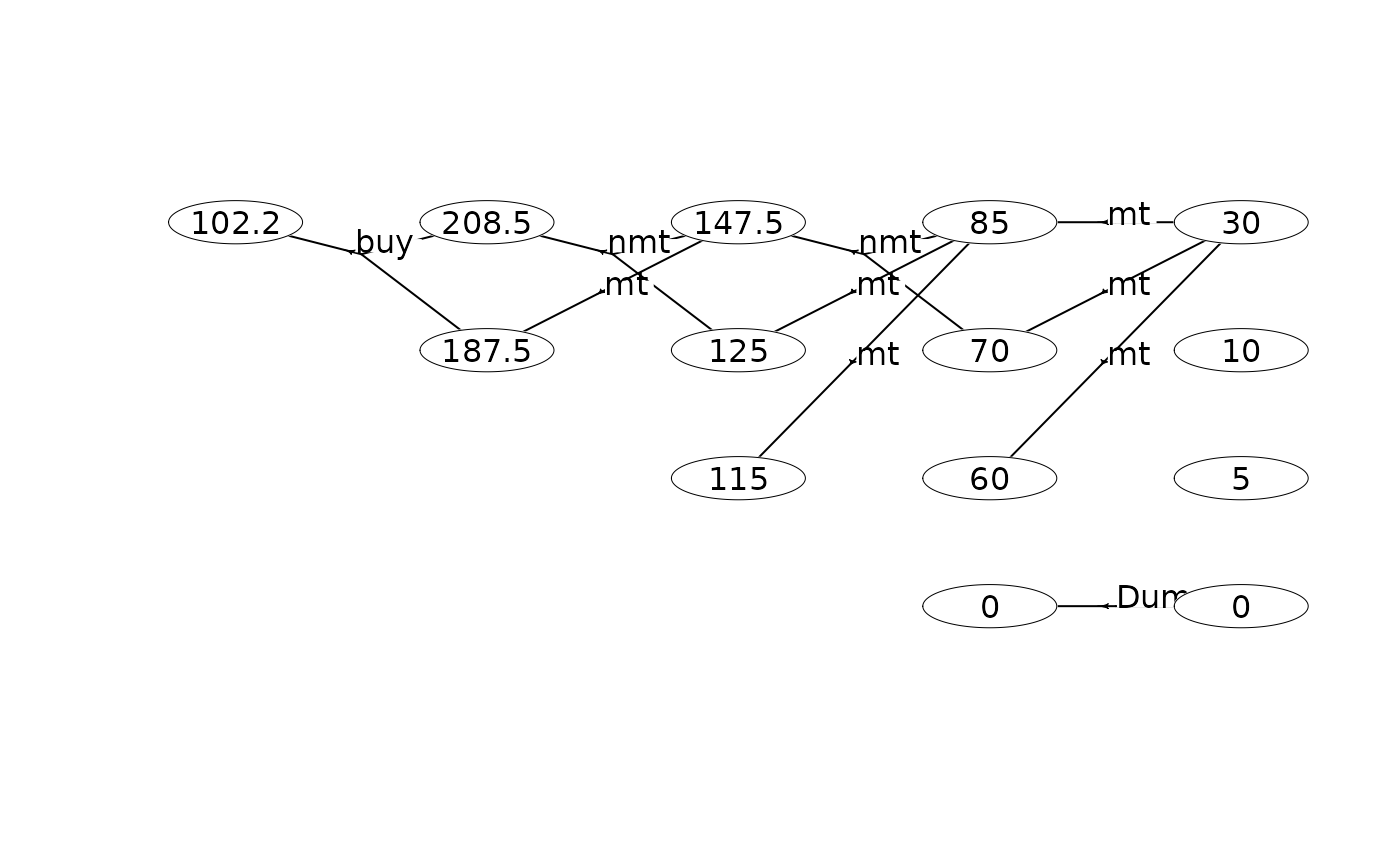
 scrapValues <- c(30, 10, 5, 0) # scrap values (the values of the 4 states at stage 4)
runValueIte(mdp, "Net reward" , termValues = scrapValues)
#> Run value iteration with epsilon = 0 at most 1 time(s)
#> using quantity 'Net reward' under reward criterion.
#> Finished. Cpu time 9.001e-06 sec.
plot(mdp, hyperarcColor = "policy") # highlight optimal policy
#> Joining with `by = join_by(sId, aIdx)`
scrapValues <- c(30, 10, 5, 0) # scrap values (the values of the 4 states at stage 4)
runValueIte(mdp, "Net reward" , termValues = scrapValues)
#> Run value iteration with epsilon = 0 at most 1 time(s)
#> using quantity 'Net reward' under reward criterion.
#> Finished. Cpu time 9.001e-06 sec.
plot(mdp, hyperarcColor = "policy") # highlight optimal policy
#> Joining with `by = join_by(sId, aIdx)`
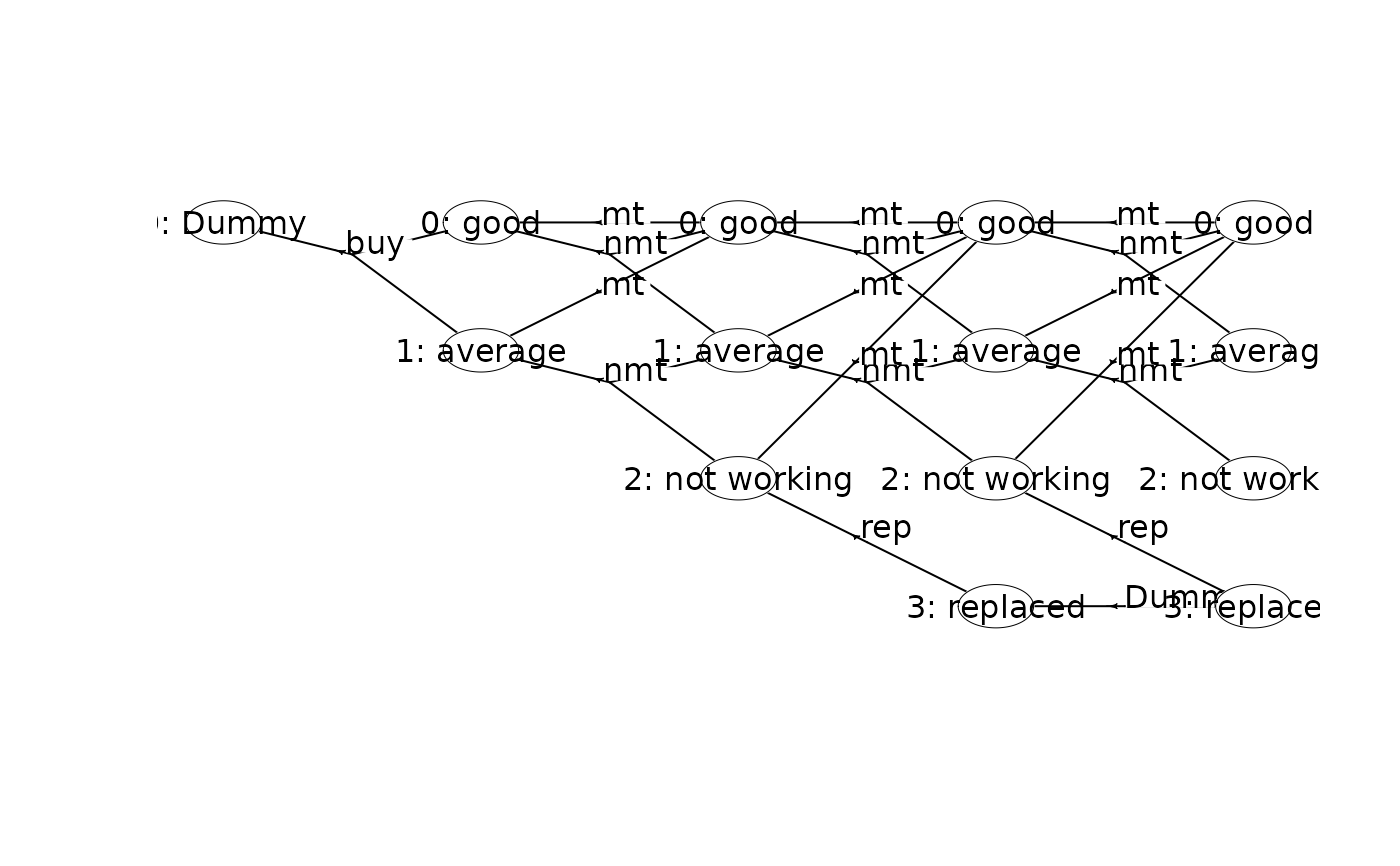
 plot(mdp, hyperarcShow = "policy", nodeLabel = "weight") # show only optimal policy
plot(mdp, hyperarcShow = "policy", nodeLabel = "weight") # show only optimal policy
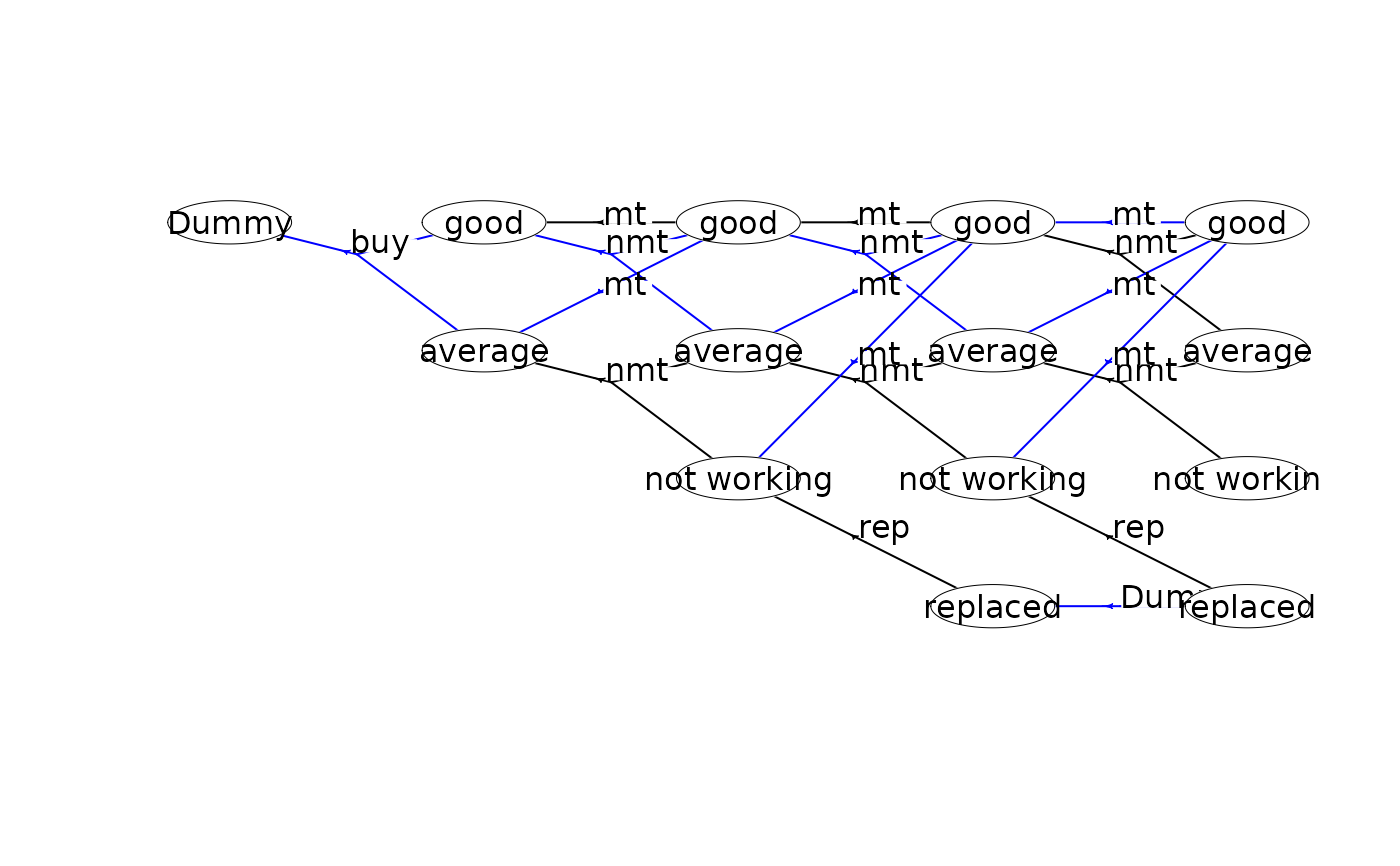
 #### An infinite-horizon maintenance problem ####
mdp<-loadMDP("hct611-1_")
#> Read binary files (0.000128403 sec.)
#> Build the HMDP (2.97e-05 sec.)
#> Checking MDP and found no errors (1.5e-06 sec.)
plot(mdp) # plot the first two stages
#### An infinite-horizon maintenance problem ####
mdp<-loadMDP("hct611-1_")
#> Read binary files (0.000128403 sec.)
#> Build the HMDP (2.97e-05 sec.)
#> Checking MDP and found no errors (1.5e-06 sec.)
plot(mdp) # plot the first two stages
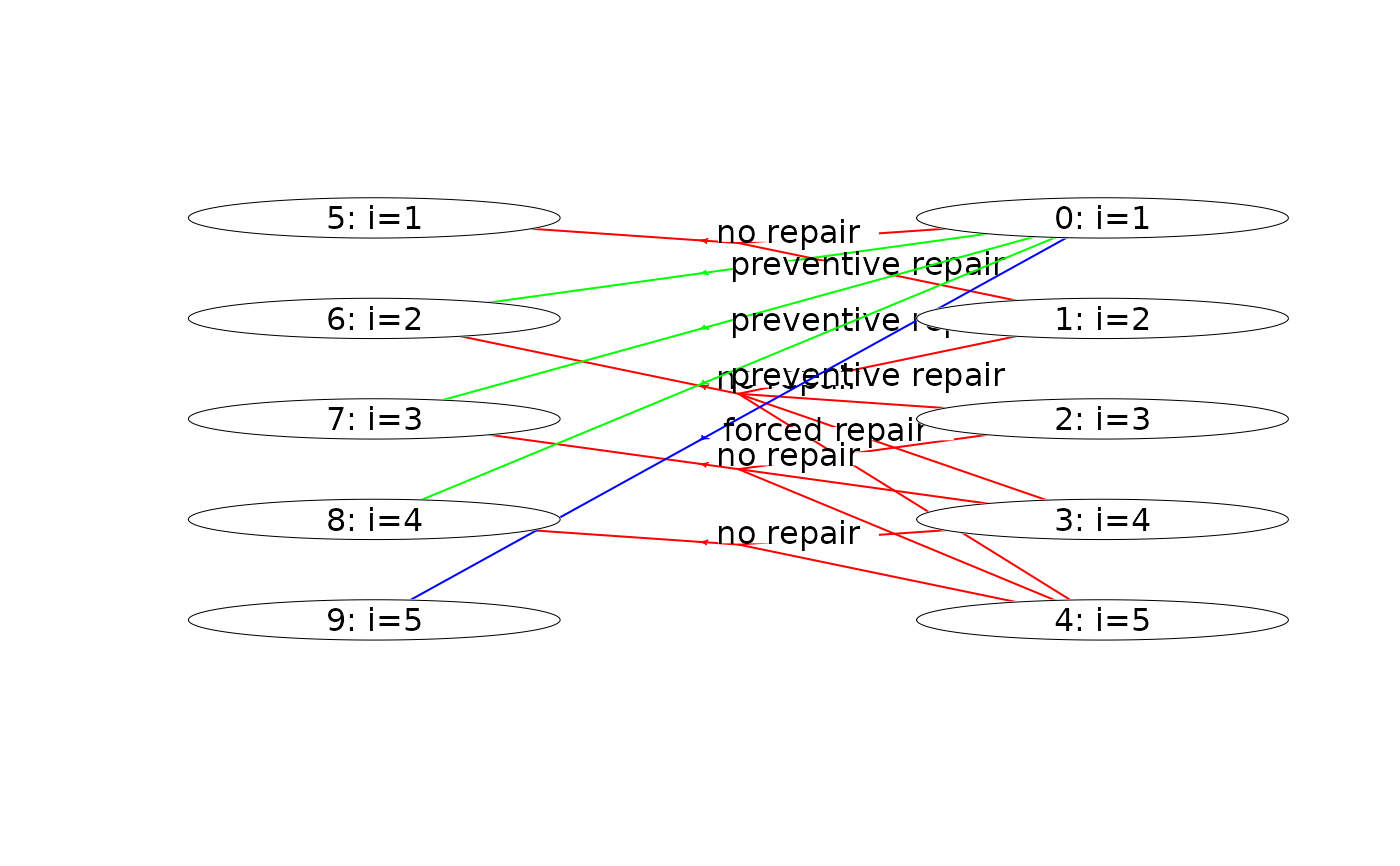
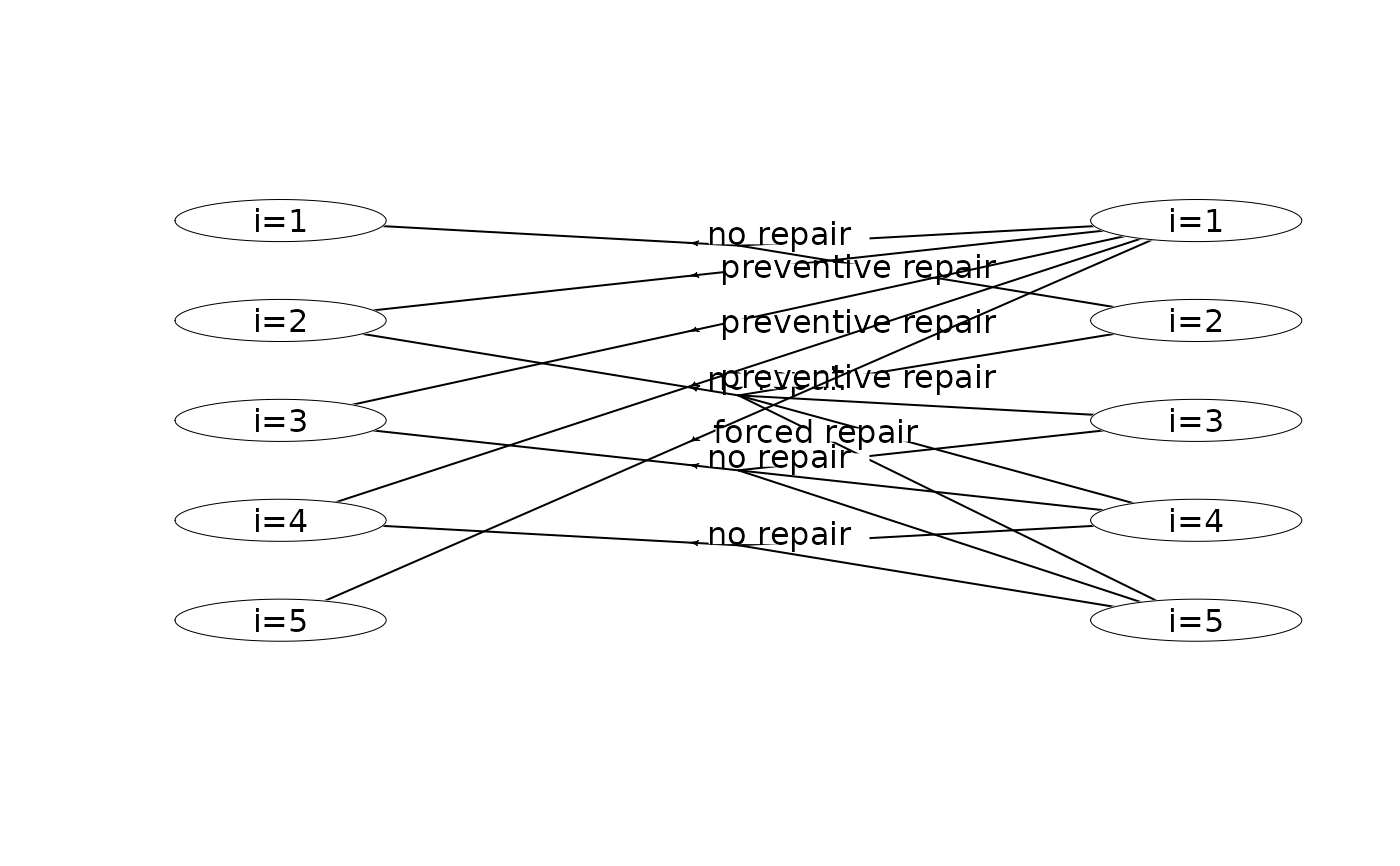 plot(mdp, hyperarcColor = "label") # colors based on labels
plot(mdp, hyperarcColor = "label") # colors based on labels
 plot(mdp, hyperarcColor = "label", nodeLabel = "sId:label") # node labels are 'sId: label'
plot(mdp, hyperarcColor = "label", nodeLabel = "sId:label") # node labels are 'sId: label'
 runPolicyIteAve(mdp,"Net reward","Duration")
#> Run policy iteration under average reward criterion using
#> reward 'Net reward' over 'Duration'. Iterations (g):
#> 1 (-0.512821) 2 (-0.446154) 3 (-0.43379) 4 (-0.43379) finished. Cpu time: 1.5e-06 sec.
#> [1] -0.43379
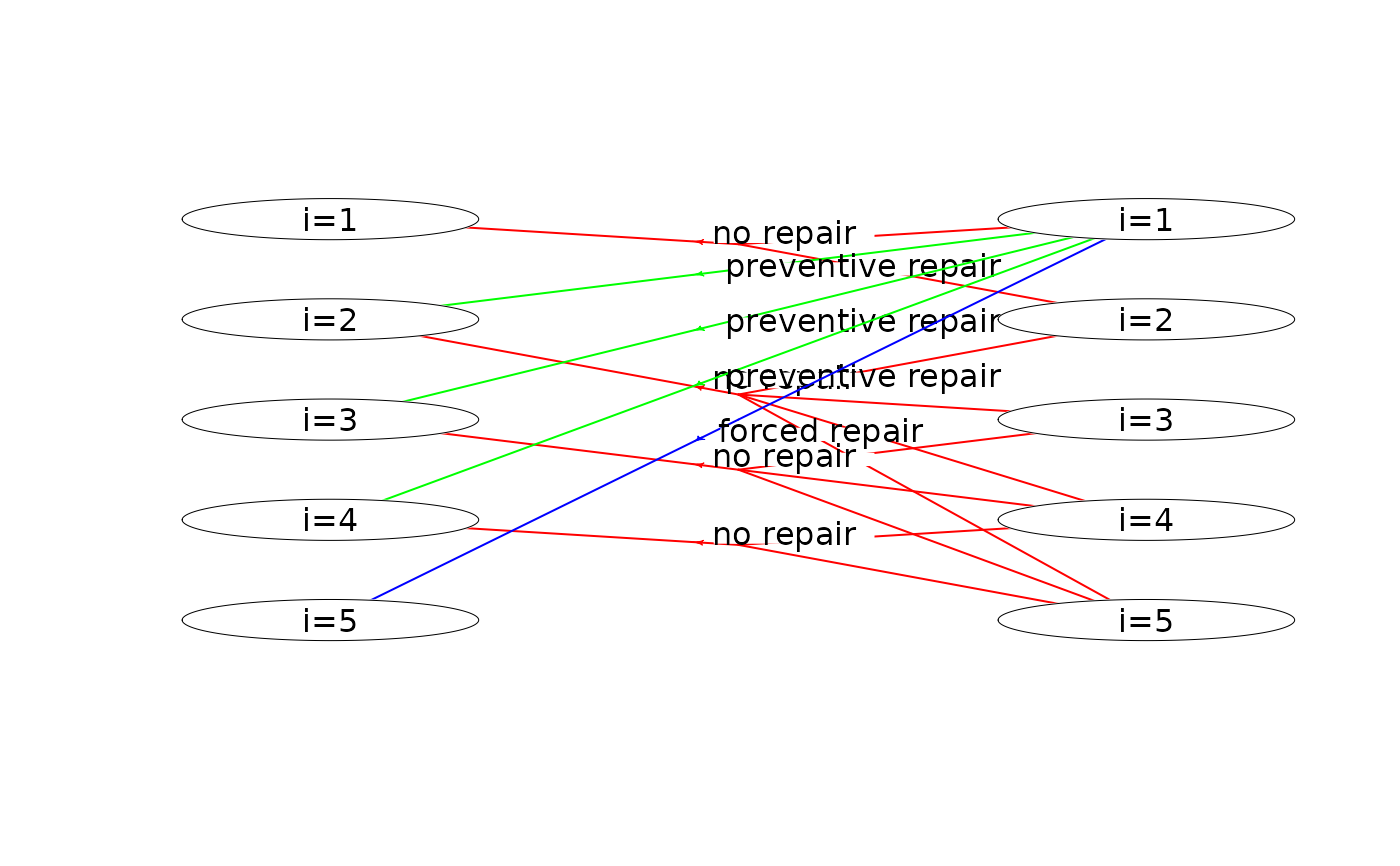
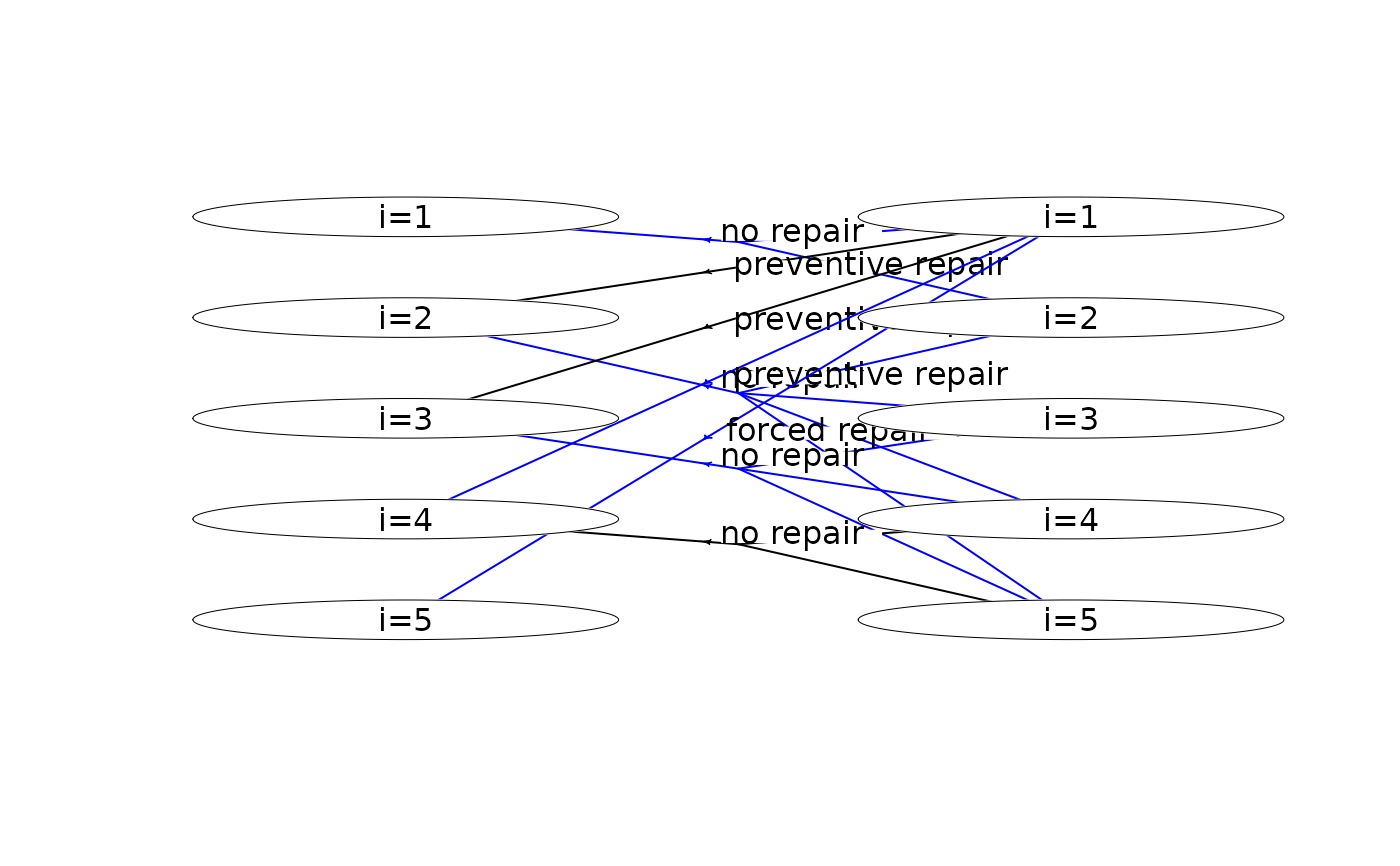
plot(mdp, hyperarcColor = "policy") # highlight optimal policy
#> Joining with `by = join_by(sId, aIdx)`
runPolicyIteAve(mdp,"Net reward","Duration")
#> Run policy iteration under average reward criterion using
#> reward 'Net reward' over 'Duration'. Iterations (g):
#> 1 (-0.512821) 2 (-0.446154) 3 (-0.43379) 4 (-0.43379) finished. Cpu time: 1.5e-06 sec.
#> [1] -0.43379
plot(mdp, hyperarcColor = "policy") # highlight optimal policy
#> Joining with `by = join_by(sId, aIdx)`
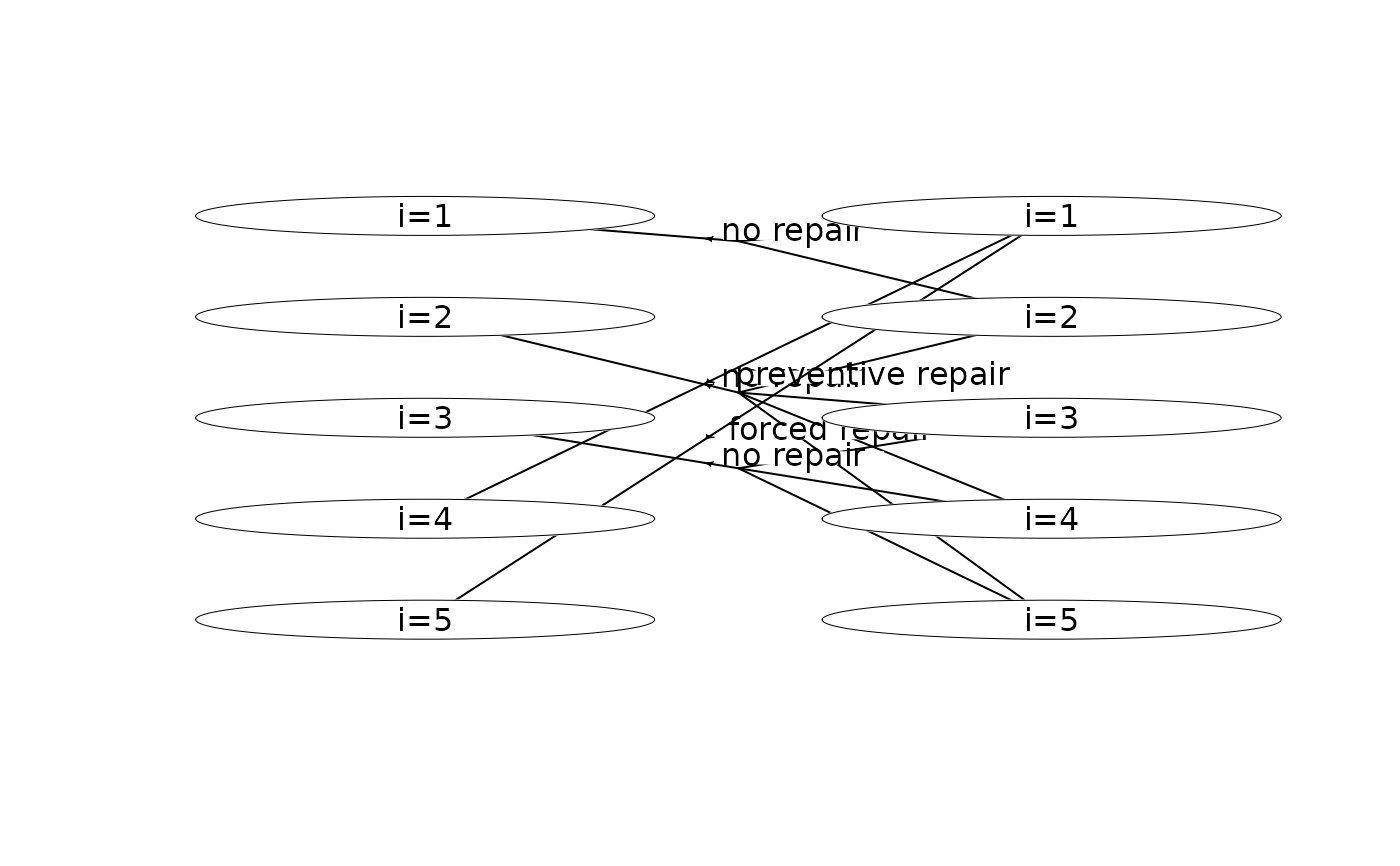
 plot(mdp, hyperarcShow = "policy") # show only optimal policy
plot(mdp, hyperarcShow = "policy") # show only optimal policy
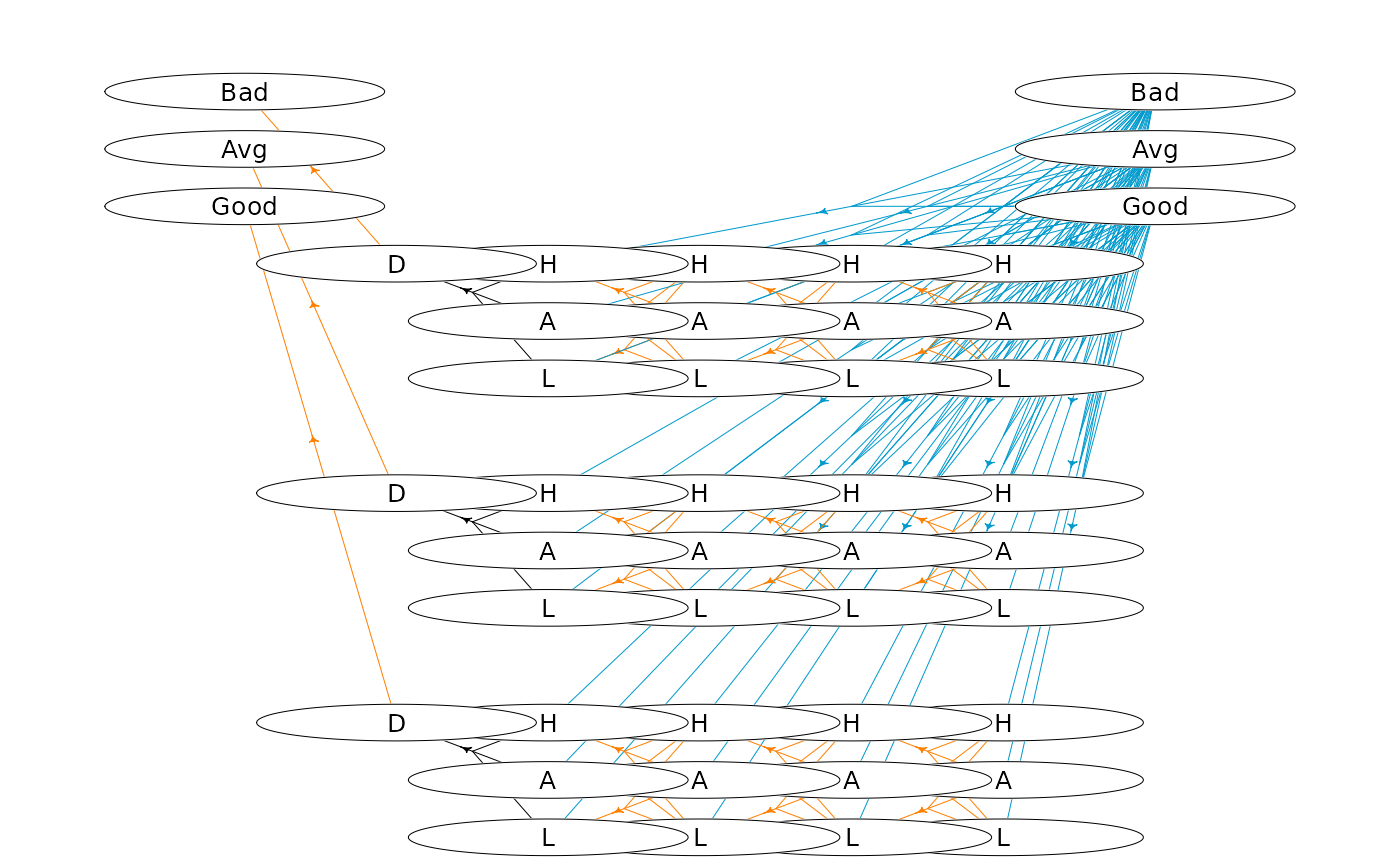
 #### An infinite-horizon hierarchical replacement problem ####
library(magrittr)
mdp<-loadMDP("cow_")
#> Read binary files (0.000247506 sec.)
#> Build the HMDP (0.000201505 sec.)
#> Checking MDP and found no errors (4.5e-06 sec.)
hgf <- getHypergraph(mdp)
# modify labels
dat <- hgf$nodes %>%
dplyr::mutate(label = dplyr::case_when(
label == "Low yield" ~ "L",
label == "Avg yield" ~ "A",
label == "High yield" ~ "H",
label == "Dummy" ~ "D",
label == "Bad genetic level" ~ "Bad",
label == "Avg genetic level" ~ "Avg",
label == "Good genetic level" ~ "Good",
TRUE ~ "Error"
))
# assign nodes to grid ids
dat$gId[1:3]<-85:87
dat$gId[43:45]<-1:3
getGId<-function(process,stage,state) {
if (process==0) start=18
if (process==1) start=22
if (process==2) start=26
return(start + 14 * stage + state)
}
idx<-43
for (process in 0:2)
for (stage in 0:4)
for (state in 0:2) {
if (stage==0 & state>0) break
idx<-idx-1
#cat(idx,process,stage,state,getGId(process,stage,state),"\n")
dat$gId[idx]<-getGId(process,stage,state)
}
hgf$nodes <- dat
# modify labels
dat <- hgf$hyperarcs %>%
dplyr::mutate(label = dplyr::case_when(
label == "Replace" ~ "R",
label == "Keep" ~ "K",
label == "Dummy" ~ "D",
TRUE ~ "Error"
),
col = dplyr::case_when(
label == "R" ~ "deepskyblue3",
label == "K" ~ "darkorange1",
label == "D" ~ "black",
TRUE ~ "Error"
),
lwd = 0.5,
label = ""
)
hgf$hyperarcs <- dat
# plot hypergraph
oldpar <- par(mai = c(0, 0, 0, 0))
plotHypergraph(gridDim = c(14, 7), hgf, cex = 0.8, radx = 0.02, rady = 0.03)
#### An infinite-horizon hierarchical replacement problem ####
library(magrittr)
mdp<-loadMDP("cow_")
#> Read binary files (0.000247506 sec.)
#> Build the HMDP (0.000201505 sec.)
#> Checking MDP and found no errors (4.5e-06 sec.)
hgf <- getHypergraph(mdp)
# modify labels
dat <- hgf$nodes %>%
dplyr::mutate(label = dplyr::case_when(
label == "Low yield" ~ "L",
label == "Avg yield" ~ "A",
label == "High yield" ~ "H",
label == "Dummy" ~ "D",
label == "Bad genetic level" ~ "Bad",
label == "Avg genetic level" ~ "Avg",
label == "Good genetic level" ~ "Good",
TRUE ~ "Error"
))
# assign nodes to grid ids
dat$gId[1:3]<-85:87
dat$gId[43:45]<-1:3
getGId<-function(process,stage,state) {
if (process==0) start=18
if (process==1) start=22
if (process==2) start=26
return(start + 14 * stage + state)
}
idx<-43
for (process in 0:2)
for (stage in 0:4)
for (state in 0:2) {
if (stage==0 & state>0) break
idx<-idx-1
#cat(idx,process,stage,state,getGId(process,stage,state),"\n")
dat$gId[idx]<-getGId(process,stage,state)
}
hgf$nodes <- dat
# modify labels
dat <- hgf$hyperarcs %>%
dplyr::mutate(label = dplyr::case_when(
label == "Replace" ~ "R",
label == "Keep" ~ "K",
label == "Dummy" ~ "D",
TRUE ~ "Error"
),
col = dplyr::case_when(
label == "R" ~ "deepskyblue3",
label == "K" ~ "darkorange1",
label == "D" ~ "black",
TRUE ~ "Error"
),
lwd = 0.5,
label = ""
)
hgf$hyperarcs <- dat
# plot hypergraph
oldpar <- par(mai = c(0, 0, 0, 0))
plotHypergraph(gridDim = c(14, 7), hgf, cex = 0.8, radx = 0.02, rady = 0.03)
 par(oldpar)
## Reset working dir
setwd(wd)
par(oldpar)
## Reset working dir
setwd(wd)